Training a Stanford NER Model in Python
Stanford NER is a good implementation of a Named Entity Recognizer (NER) using Conditional Random Fields (CRFs). CRFs are no longer near state of the art for NER, having been overtaken by LSTM-CRFs, which have since been overtaken by Transformer models. However CRFs are still a reasonable baseline, and Stanford NER is used in many papers which is good for reproducibility. It doesn’t have a Python binding (a CRF library that does is CRFsuite), but with some work we can train and test a model in Python.
I replicated the benchmark in A Named Entity Based Approach to Model Recipes, by Diwan, Batra, and Bagler using Stanford NER, and check it using seqeval. Evaluating NER is surprisingly tricky, as David Batista explains, and I want to check that the results in the paper are the same as what seqeval gives, so that I’m giving a fair comparison to other models. Thanks to the authors sharing the training data on github I was able to do this, as you can see in the Jupyter Notebook.
The rest of this article goes through how to train and evaluate a Stanford NER model using Python, and that the scores output by Stanford NLP on the test set match those produced by seqeval.
Setting up Stanford NLP
The stanza library has both great neural network based models for linguistic analysis (see my previous writeup), but also an interface to Stanford Core NLP. Unfortunately it doesn’t provide a direct way of training an NER model using Core NLP, however we can do it ourselves using the stanford-corenlp JAR it installs. The first step is to install the models and find the path to the Core NLP JAR.
import os
from pathlib import Path
import stanza
stanza.install_corenlp()
# Reimplement the logic to find the path where stanza_corenlp is installed.
core_nlp_path = os.getenv('CORENLP_HOME', str(Path.home() / 'stanza_corenlp'))
# A heuristic to find the right jar file
classpath = [str(p) for p in Path(core_nlp_path).iterdir()
if re.match(r"stanford-corenlp-[0-9.]+\.jar", p.name)][0]Train NER Model
The Stanford NER model requires data where each line is a token, followed by a tab, followed by the NER tag. A blank line represents a sentence break. In this case I could get the relevant training and test repository in this format already.
The NER model has to be configured; but there’s no information on the paper on what features and hyperparameters are used. I copied the template configuration out of the FAQ, which happened to work well. The template can be saved to a file and then referred to when training.
def ner_prop_str(train_files: List[str],
test_files: List[str],
output: str) -> str:
"""Returns configuration string to train NER model"""
train_file_str = ','.join(train_files)
test_file_str = ','.join(test_files)
return f"""
trainFileList = {train_file_str}
testFiles = {test_file_str}
serializeTo = {output}
map = word=0,answer=1
useClassFeature=true
useWord=true
useNGrams=true
noMidNGrams=true
maxNGramLeng=6
usePrev=true
useNext=true
useSequences=true
usePrevSequences=true
maxLeft=1
useTypeSeqs=true
useTypeSeqs2=true
useTypeySequences=true
wordShape=chris2useLC
useDisjunctive=true
"""For more information on the parameters you can check the NERFeatureFactory documentation or the source. We need to write this to a file so we can write a wrapper to do this
def write_ner_prop_file(ner_prop_file: str,
train_files: List[str],
test_files: List[str],
output_file: str) -> None:
with open(ner_prop_file, 'wt') as f:
props = ner_prop_str(train_files, test_files, output_file)
f.write(props)The actual NER Training process is in Java, so we’ll run a Java process to train a model and return the path to the model file. We’ll also print out the report from stderr summarising the training.
import subprocess
from typing import List
def train_model(model_name: str,
train_files: List[str],
test_files: List[str],
print_report: bool = True,
classpath: str = classpath) -> str:
"""Trains CRF NER Model using StanfordNLP"""
model_file = f'{model_name}.model.ser.gz'
ner_prop_filename = f'{model_name}.model.props'
write_ner_prop_file(ner_prop_filename, train_files, test_files, model_file)
result = subprocess.run(
['java',
'-Xmx2g',
'-cp', classpath,
'edu.stanford.nlp.ie.crf.CRFClassifier',
'-prop', ner_prop_filename],
capture_output=True,
check=True)
if print_report:
print(*result.stderr.decode('utf-8').split('\n')[-11:], sep='\n')
return model_fileRunning training on the AllRecipes.com train and test set produced an output like this.
The summary report shows for each model and entity type:
- True Positives (TP): The number of times that entity was predicted correctly
- False Positives (FP): The number of times that entity in the text but not predicted correctly
- False Negative (FN): The number of times that entity was not in the text and predicted
- Precision (P): Probability a predicted entity is correct, TP/(TP+FP)
- Recall (R): Probability a correct entity is predicted, TP/(TP+FN)
- F1 Score (F1): Harmonic mean of precision and recall, 2/(1/P + 1/R).
CRFClassifier tagged 2788 words in 483 documents at 9992.83 words per second.
Entity P R F1 TP FP FN
DF 1.0000 0.9608 0.9800 49 0 2
NAME 0.9297 0.9279 0.9288 463 35 36
QUANTITY 1.0000 0.9962 0.9981 522 0 2
SIZE 1.0000 1.0000 1.0000 20 0 0
STATE 0.9601 0.9633 0.9617 289 12 11
TEMP 0.8750 0.7000 0.7778 7 1 3
UNIT 0.9819 0.9841 0.9830 434 8 7
Totals 0.9696 0.9669 0.9682 1784 56 61The Totals F1 score of 0.9682 exactly matched what was reported in the paper. Now let’s try to manually evaluate the test set using seqeval.
Running the model
Stanza has a robust way of running CoreNLP and annotating texts, as per the documentation. We can configure the NER model used to the one that we just trained. Because the text we’ve used is pre-tokenized I’m just going to join them with a space and tokenize on whitespace; for ingredients we want quantities like 1/2 to be treated as a single token but the default tokenizer will split them. When I first ran the annotations it would sometime output NUMBER, which wasn’t an input entity; it turns out this is hardcoded and we have to diable the numeric classifiers.
from stanza.server import CoreNLPClient
def annotate_ner(ner_model_file: str,
texts: List[str],
tokenize_whitespace: bool = True):
properties = {"ner.model": ner_model_file,
"tokenize.whitespace": tokenize_whitespace,
"ner.applyNumericClassifiers": False}
annotated = []
with CoreNLPClient(
annotators=['tokenize','ssplit','ner'],
properties=properties,
timeout=30000,
be_quiet=True,
memory='6G') as client:
for text in texts:
annotated.append(client.annotate(text))
return annotatedThe annotated data will have many attributes, but we’re just interested in the input words and named entities so we’ll extract them into a dictionary. Note that we extract the coarseNER; sometimes another default NER model predicts a fine grained NER (like NATIONALITY) which writes into the ner attribute if it’s empty. Using coarseNER means we only get tags from our training set.
def extract_ner_data(annotation) -> Dict[str, List[str]]:
tokens = [token for sentence in annotation.sentence
for token in sentence.token]
return {'tokens': [t.word for t in tokens],
'ner': [t.coarseNER for t in tokens]}
def ner_extract(ner_model_file: str,
texts: List[str],
tokenize_whitespace: bool = True) -> List[Dict[str, List[str]]]:
annotations = annotate_ner(ner_model_file, texts, tokenize_whitespace)
return [extract_ner_data(ann) for ann in annotations]Now if we’ve got the test tokens as a list containing lists of words, and the test labels as a list containing corresponding lists of NER tags we can run them through the model.
test_texts = [' '.join(text) for text in test_tokens]
pred_labels = [text['ner'] for text in ner_extract(modelfile, texts)]Evaluating with seqeval
The library seqeval provides robust sequence labelling metrics. In particular scores should be at an entity level; you don’t get it right unless you predict exactly the tokens in an entity. I wanted to check seqeval gave similar results to the sumamary report above.
Seqeval expects the tags to be in one of the standard tagging formats, but the data I had just had labels (like NAME, QUANTITY, and UNIT). It is impossible to disambiguate adjacent tags of the same entity type, but the annotations mostly assume there can only be one of each kind of entity in an ingredient. The simplest way to convert it is into IOB-1, which only adds a B tag when there are two adjacent tags of the same entity type. Since we’re assuming this doesn’t happen we just need to prepend I- to all tags other than O.
def convert_to_iob1(tokens):
return ['I-' + label if label != 'O' else 'O' for label in tokens]
result = convert_to_iob1(['QUANTITY', 'SIZE', 'NAME', 'NAME', 'O', 'STATE'])
expected = ['I-QUANTITY', 'I-SIZE', 'I-NAME', 'I-NAME', 'O', 'I-STATE']
assert result == expectedThen we can get the classification report using seqeval
from seqeval.metrics import classification_report
actual_labels = [convert_to_iob1(text) for text in actual_labels]
pred_labels = [convert_to_iob1(text) for text in pred_labels]
print(classification_report(actual_labels, pred_labels, digits=4))The output report matches the report from Stanford NLP precisely. Note that it uses the support (total number of actual entities) instead of the True Positives, False Positives, and False Negatives, but actually they are equivalent.
- support = TP + FN
- TP = R * support
- FP = TP (1/P - 1)
- FN = support - TP
precision recall f1-score support
DF 1.0000 0.9608 0.9800 51
NAME 0.9297 0.9279 0.9288 499
QUANTITY 1.0000 0.9962 0.9981 524
SIZE 1.0000 1.0000 1.0000 20
STATE 0.9601 0.9633 0.9617 300
TEMP 0.8750 0.7000 0.7778 10
UNIT 0.9819 0.9841 0.9830 441
micro avg 0.9696 0.9669 0.9682 1845
macro avg 0.9638 0.9332 0.9471 1845
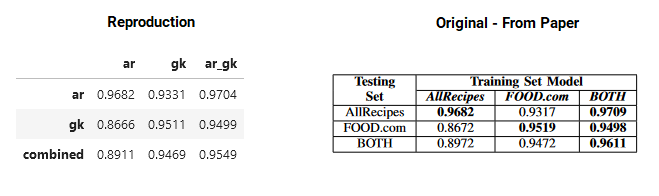
weighted avg 0.9695 0.9669 0.9682 1845Using seqeval on all the training and test sets in the paper I could reproduce their f1-scores within 0.01. Below shows my results (left) and the results on the paper (right).
The Stanford NER library is a bit under-documented and has some surprising features, but with some work we can get it to run in Python. The metrics it produces line up with those from seqeval.
